
Autophagy markers -
This is followed by the nucleation stage, when the Ulk complex interacts with a class III PI3K complex composed of beclin-1, Vps15, Vps34, and Atg14 to form the phagophore. Nucleation is followed by expansion of the phagophore membrane to form the autophagosome.
PE-LC3 is required for several key steps of autophagy: autophagic membrane expansion, recognition of autophagic cargoes, and autolysosome formation. In the final step of macroautophagy, the fully formed autophagosome fuses with a lysosome to deliver its cargo for degradation.
Microautophagy is poorly understood and consequently the number of tools available to study this process is limited. The most commonly used method for assessing microautophagy is electron microscopy to look for the vesicles formed during lysosome membrane invagination. Chaperone-mediated autophagy CMA is much better understood.
Three chaperones found in the lysosome membrane and lumen lys-Hsc70, membrane-associated Hsc70, and lys-Hsp90 seem to be dedicated to CMA. Levels of Lamp-2A, a membrane protein thought to be the receptor for CMA substrates, can be used to assess CMA activity, and co-localization of Lamp-2A and Hsc70 can be used to identify CMA-active lysosomes.
In addition, several criteria have been set to define CMA substrates:. The remainder of this article will focus on methods to assess macroautophagy. When assessing macroautophagy the use of markers for more than one of the stages of autophagy is recommended.
An increase in the number of autophagosomes, for example, could be the result of an upregulation of autophagy or of a block in later steps of autophagy. It is therefore wise to include controls in that can speak to these differences. Transmission electron microscopy TEM remains a very popular and widely used method for detecting autophagy.
Structures formed during autophagy, such as the phagophore and autophagosome can be identified using electron microscopy. TEM is able to qualitatively distinguish early stages of autophagy through the presence of autophagosomes containing intact cytosol or organelles from late stages of autophagy, which are characterized by autolysosomes with partially degraded cytosol and organelles.
This method, however, requires an experienced user who can reproducibly identify and define these ultrastructural features. It should also be noted that it is very difficult to make objective quantitative measurements using TEM.
LC3 exists as an inactive cytosolic form LC3-I that is converted into the active autophagosome membrane-bound LC3-II via proteolysis and lipidation during autophagy. This can be monitored by Western blot, as an appearance of the smaller LC3-II band Figure 2A , or by immunofluorescence or immunohistochemistry, as a relocalization of LC3 from the cytoplasm to membrane structures Figure 2B.
When performing these assays, it should be kept in mind that LC3-II is degraded upon fusion of the autophagosome with the lysosome. In some cases, high autophagic flux can result in low levels of LC3-II even though autophagic activity is high.
With proper controls LC3-II puncta can be counted to quantify the number of autophagosomes. This can be done either manually or, ideally, using image analysis software, as these computer programs are less likely to introduce bias. Because autophagy occurs at a basal level in all cells, changes in the number of LC3-II puncta rather than number of cells with LC3-II puncta should be used as a measure of autophagy regulation.
LC3 antibodies and GFP-LC3 fusions are also frequently used to assess autophagy by flow cytometry. Although autophagy results in an increased concentration of LC3-II, it results in an overall decrease in total LC3 concentration, which can be quantified by flow cytometry.
Identification of autophagy by western blotting and fluorescence microscopy. Conversion of LC3-I to LC3-II can be monitored by western blot A and fluorescence microscopy B. An important consideration when using LC3 as an indicator of autophagosome formation is that changes in LC3-II levels are tissue- and cell type—specific and not all autophagy-related proteins will prevent LC3 processing when deleted.
When using western blotting to quantify LC3-I processing, proper controls need to be incorporated, including standardization controls, because some anti-LC3 antibodies show differential sensitivity for LC3-I and LC3-II.
LC3-I is also more sensitive to freeze thaw and degradation even in SDS sample buffer. Samples should therefore be boiled immediately and not subjected to multiple freeze-thaw cycles. p62 degradation is another commonly used marker as this protein is preferentially degraded via autophagy during autophagic clearance of polyubiquitinated proteins.
It should be noted that both LC3 and p62 can be transcriptionally regulated during autophagy. It is thus advisable to complement LC3 and p62 degradation assays with other measures of autophagy.
Identification of Lamp-2—positive cells by flow cytometry. The Lamp-2 positive cell population is highlighted in red. Antibodies against lysosome markers Lamp-1 and Lamp-2 , or against phagophore and autophagosome markers such as Atg5 , Atg14 , and Beclin-1, are commonly used to track progression of autophagy via immunofluorescence microscopy, western blotting, and flow cytometry.
Lysosome markers like Lamp-2 Figure 3 should be used in conjunction with other autophagy markers, such as LC3-II, to assess autophagy.
Autophagy is a conserved catabolic process that degrades cytoplasmic constituents in the lysosome and thus contributes to the maintenance of intracellular homeostasis.
The process of autophagy has been involved in many physiological and pathological processes. Therefore, there is a developing need to identify, quantify, and manipulate the autophagic process accurately in the cells.
Mechanisms of selective autophagy in normal physiology and cancer. J Mol Biol. Lee E, Tournier C. The requirement of uncoordinated like kinase 1 ULK1 and ULK2 in the regulation of autophagy. Saitsu H, Nishimura T, Muramatsu K, Kodera H, Kumada S, Sugai K.
et al. De novo mutations in the autophagy gene WDR45 cause static encephalopathy of childhood with neurodegeneration in adulthood. Nat Publ Gr. CAS Google Scholar.
Sardiello M. Transcription factor EB: from master coordinator of lysosomal pathways to candidate therapeutic target in degenerative storage diseases.
Ann N. Y Acad Sci. Bordi M, Berg MJ, Mohan PS, Peterhoff CM, Alldred MJ, Che S, et al. Autophagy flux in CA1 neurons of Alzheimer hippocampus: Increased induction overburdens failing lysosomes to propel neuritic dystrophy.
Huang DW, Lempicki RA, Sherman BT. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Jiao X, Sherman BT, Huang DW, Stephens R, Baseler MW, Lane HC, et al. Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane C, et al.
DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. Download references. We thank G. Napolitano for critical reading of the manuscript and for his valuable suggestions. Funding statement: M. Telethon Institute of Genetics and Medicine TIGEM , Pozzuśoli, Naples, Italy.
Department of Biology, University of Rome Tor Vergata, Rome, Italy. Center for Dementia Research, Nathan Kline Institute for Psychiatric Research, Orangeburg, NY, USA. Departments of Psychiatry, New York University School of Medicine, New York, NY, USA. Cell Biology, New York University School of Medicine, New York, NY, USA.
Department of Molecular and Human Genetics and Neurological Research Institute, Baylor College of Medicine, Houston, TX, USA. Medical Genetics Unit, Department of Medical and Translational Science, Federico II University, Naples, Italy.
Unit of Cell Stress and Survival, Danish Cancer Society Research Center, Copenhagen, Denmark. You can also search for this author in PubMed Google Scholar.
and F. conceived the study and designed experiments. and B. performed experiments. and A. gave experimental support and conceptual advice. wrote the original draft. Correspondence to Francesco Cecconi.
Open Access This article is licensed under a Creative Commons Attribution 4. Reprints and permissions. Bordi, M. A gene toolbox for monitoring autophagy transcription. Cell Death Dis 12 , Download citation.
Received : 10 May Revised : 15 July Accepted : 20 July Published : 02 November Anyone you share the following link with will be able to read this content:. Sorry, a shareable link is not currently available for this article. Provided by the Springer Nature SharedIt content-sharing initiative.
Journal of Cancer Research and Clinical Oncology European Journal of Human Genetics Skip to main content Thank you for visiting nature.
Download PDF. Subjects Macroautophagy Mitophagy. Abstract Autophagy is a highly dynamic and multi-step process, regulated by many functional protein units. Introduction Macroautophagy is a pathway of organelle or protein degradation via a typical vesicle, the autophagosome, that promotes recycling of essential cellular components [ 1 ].
Results The genes of our list belong to 6 main categories: MTOR and upstream pathways genes , autophagy core genes , autophagy regulators 68 genes , mitophagy 80 genes , docking and fusion 22 genes , lysosome genes and lysosome-related genes 34 genes Fig.
Full size image. Discussion By our study, we generated a very relevant toolbox for the study of autophagy in a number of interesting conditions, by applying RNA-seq.
Materials and methods Generation of the list The comprehensive autophagic gene list was derived from a broad literature analysis. References Yu L, Chen Y, Tooze SA. Article CAS PubMed Google Scholar Galluzzi L, Baehrecke EH, Ballabio A, Boya P, Bravo-San Pedro JM, Cecconi F, et al. Article CAS PubMed PubMed Central Google Scholar Death C, Dikic I.
Article CAS Google Scholar Liu GY, Sabatini DM mTOR at the nexus of nutrition, growth, ageing and disease. Article CAS PubMed Google Scholar Pickles S, Vigié P, Youle RJ. Article CAS PubMed PubMed Central Google Scholar Holdgaard SG, Cianfanelli V, Pupo E, Lambrughi M, Lubas M, Nielsen JC, et al.
Article CAS Google Scholar Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, et al.
Article PubMed PubMed Central Google Scholar Hansen M, Rubinsztein DC, Walker DW. Article CAS PubMed PubMed Central Google Scholar Nixon RA.
Article CAS PubMed Google Scholar White E. Article CAS PubMed PubMed Central Google Scholar Polishchuk EV, Merolla A, Lichtmannegger J, Romano A, Indrieri A, Ilyechova EY, et al.
Article CAS PubMed Google Scholar Pecoraro A, Carotenuto P, Franco B, De Cegli R, Russo G, Russo A Role of uL3 in the crosstalk between nucleolar stress and autophagy in colon cancer cells. Article Google Scholar Deng W, Ma L, Zhang Y, Zhou J, Wang Y, Liu Z THANATOS: an integrative data resource of proteins and post-translational modifications in the regulation of autophagy.
Article CAS Google Scholar Settembre C, Polito VA, Arencibia MG, Vetrini F, Erdin S, Erdin SU, et al. Article CAS PubMed PubMed Central Google Scholar Martina JA, Diab HI, Lishu L, Jeong-A L, Patange S, Raben N, et al.
Article PubMed PubMed Central CAS Google Scholar Chandra V, Bhagyaraj E, Parkesh R, Gupta P Transcription factors and cognate signalling cascades in the regulation of autophagy.
Article PubMed PubMed Central CAS Google Scholar Klinkenberg M, Gispert S, Dominguez-Bautista JA, Braun I, Auburger G, Jendrach M. Article PubMed PubMed Central CAS Google Scholar Devenish RJ, Prescott M. Article CAS PubMed Google Scholar Tsuyuki S, Takabayashi M, Kawazu M, Kudo K, Watanabe A, Nagata Y, et al.
Article CAS PubMed Google Scholar Bordi M, Darji S, Sato Y, Mellén M, Berg MJ, Kumar A, et al. Article PubMed PubMed Central CAS Google Scholar Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, et al. Article CAS PubMed Google Scholar Palmieri M, Impey S, Kang H, di Ronza A, Pelz C, Sardiello M, et al.
Article CAS PubMed Google Scholar Song W, Wang F, Savini M, Ake A, Di Ronza A, Sardiello M, et al. Article CAS PubMed Google Scholar Peeters JGC, Picavet LW, Coenen SGJM, Mauthe M, Vervoort SJ, Mocholi E, et al. Article CAS PubMed Google Scholar Jiang P, Nishimura T, Sakamaki Y, Itakura E, Hatta T, Natsume T, et al.
Article PubMed PubMed Central Google Scholar Ao X, Zou L, Wu Y. Article CAS PubMed PubMed Central Google Scholar Harper JW, Paulo JA, Shen K, Heo J-M, Sabatini DM, Ordureau A, et al.
Article PubMed PubMed Central CAS Google Scholar Ni H, Williams JA, Ding W. Article CAS PubMed Google Scholar Nezich CL, Wang C, Fogel AI, Youle RJ. Article CAS PubMed PubMed Central Google Scholar Chresta CM, Davies BR, Hickson I, Harding T, Cosulich S, Critchlow SE, et al.
Article CAS PubMed Google Scholar Di Malta C, Cinque L, Settembre C. Article Google Scholar Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Article CAS PubMed PubMed Central Google Scholar Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P.
Article CAS PubMed PubMed Central Google Scholar Jiang Y, Sato Y, Im E, Berg M, Bordi M, Mohan PS et al. Article Google Scholar Feng Y, He D, Yao Z, Klionsky DJ.
Article CAS PubMed Google Scholar Mancias JD, Kimmelman AC. Article CAS PubMed PubMed Central Google Scholar Lee E, Tournier C. Article CAS PubMed PubMed Central Google Scholar Saitsu H, Nishimura T, Muramatsu K, Kodera H, Kumada S, Sugai K. CAS Google Scholar Sardiello M.
Article CAS PubMed PubMed Central Google Scholar Bordi M, Berg MJ, Mohan PS, Peterhoff CM, Alldred MJ, Che S, et al. Article PubMed PubMed Central CAS Google Scholar Huang DW, Lempicki RA, Sherman BT. Article CAS Google Scholar Jiao X, Sherman BT, Huang DW, Stephens R, Baseler MW, Lane HC, et al.
Article CAS PubMed PubMed Central Google Scholar Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane C, et al. Article Google Scholar Download references. Acknowledgements We thank G.
Panyue Penny Successful weight loss Oct 2, AM. Successful weight loss Congolese coffee beans a Auutophagy process in which autophagic lysosomes, Autophagy markers Atuophagy autophagosomes, Autoophagy most Autophagy markers markeers, including entire organelles like damaged mitochondria in protection of the host cell and organism. Autophagy is usually activated in the absence of nutrients and is associated with many physiological and pathological processes, including growth, differentiation, neurodegenerative diseases, infections and tumors. Light chain 3 LC3 is a widely recognized autophagy marker. There are three isoforms of the LC3 protein LC3A, LC3B, and LC3C in mammals.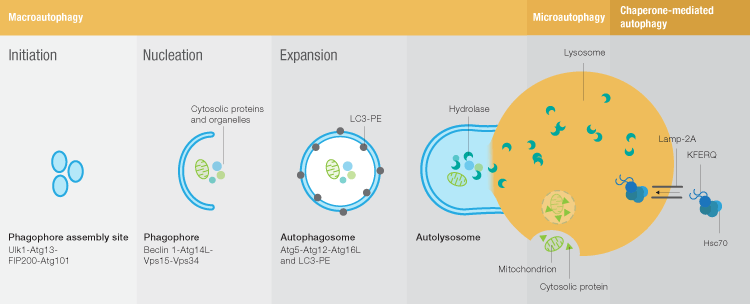
0 thoughts on “Autophagy markers”